We’re extremely excited to release APPT, the state of the art protein-protein binding affinity prediction tool. APPT performs at unprecedented speed and accuracy, outperforming existing solutions by 40% — while scanning 9,000 proteins per second.
The model is completely open source—the training code, inference code, model weights and architecture are all available on GitHub under the CC-BY-NC-SA license.
SOTA for protein–protein interactions
In order to rigorously test APPT, we used a variety of benchmarks to validate its performance, and found that it performs extremely well compared to other existing solutions.
On the Haddock benchmark, which measures models against 46 high quality protein-protein binding affinity ground truth data — APPT demonstrates significantly superior performance compared to other methods, achieving an R² value of 0.68. This represents a substantial improvement over the next best performers: PyDock (R² = 0.28), Rosetta (R² = 0.27).
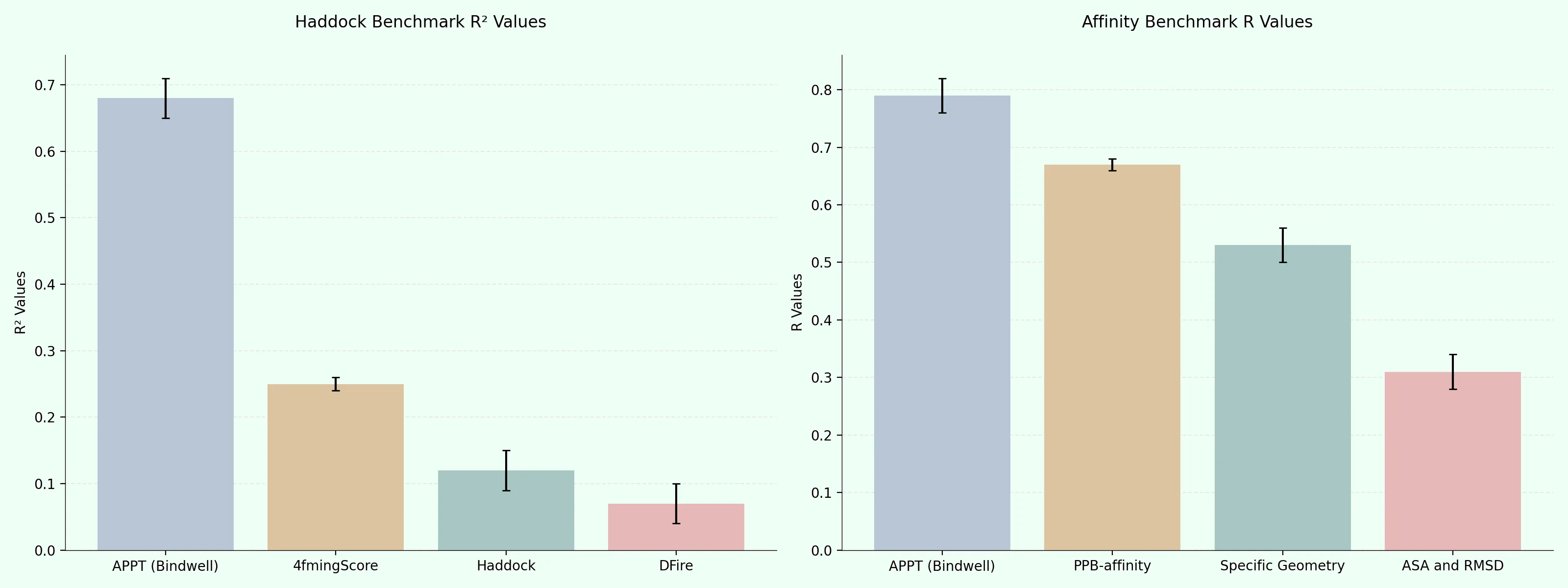
On the Affinity V5.5 benchmark dataset, our APPT model achieves a Pearson correlation coefficient (R) of over 0.8 with experimental binding affinities, setting a new state-of-the-art performance. This surpasses both:
- Existing machine learning approaches:
- PPB-Affinity and similar ML/GNN models that predict binding affinity from protein structures
- Traditional physics-based methods:
- Molecular Mechanics/Poisson-Boltzmann Surface Area (MM-PBSA)
- Molecular Mechanics/Generalized Born Surface Area (MM-GBSA)
- Methods based on specific geometric features, accessible surface area (ASA), and root-mean-square deviation (RMSD) calculations
Sequences are all you need
Many docking methods rely on empirical or semi-empirical scoring functions to estimate binding energy. These scoring functions are often over-simplified and fail to accurately capture the complex biophysics of protein-protein interactions.
Our key innovation lies in training PLMs (Protein Language Models) such as Ankh and ESM on downstream tasks using the biggest available dataset possible. For APPT, we trained the ESM2_3B model on 11,000 protein pairs — the largest sequence-affinity database yet!
The model architecture can be described as following:
- A protein embedding layer using ESM2_3B that captures both local amino acid patterns and long-range dependencies
- A transformer encoder with multi-head self-attention mechanisms
- A projection head that reduces the 2560-dimensional ESM embeddings to a more focused binding-relevant subspace
- A fully connected neural network that combines information from both proteins to estimate binding affinity
What’s next?
We’re working on improving the capabilities of APPT — refining it almost every day now! Let us know if you have any suggestions by emailing founders@bindwell.ai.